How to use covid19viz
Andree Valle
2020-09-20
how-to-use.RmdExample 1: Analyse intervention and mobility data
# library(covidPeru); library(readr)
library(covid19viz)
# library(cdcper)
library(tidyverse)
library(lubridate)
theme_set(theme_bw())
# analysis time limits --------------------------------------------------------------
min_analysis_date <- ymd(20200301)
max_analysis_date <- Sys.Date()
# covid19viz R package ----------------------------------------------------
# _import intervention data ------------------------------------------------
unesco <- read_unesco_education()
# acaps <- read_acaps_governments()
# no data in ACAPS
# acaps %>% filter(ISO3=="PER")
unesco_peru <- unesco %>%
filter(ISO=="PER") %>%
mutate(Date=dmy(Date)) %>%
group_by(Status) %>%
summarise(date_min=min(Date),
date_max=max(Date)) %>%
rename(intervention_label=Status) %>%
mutate(intervention=case_when(
intervention_label=="Closed due to COVID-19" ~ "closed",
intervention_label=="Partially open" ~ "partial"
))
# unesco_peru
# _unite intervention data -------------------------------------------------
interventions <- tibble(
date_min = ymd(20200628),
date_max = ymd(20200709),
intervention_label = "Seroprevalence study",
intervention = "seroprev"
) %>%
union_all(
unesco_peru %>%
mutate(date_max=if_else(date_max==max(date_max),
max_analysis_date,
date_max))
)
interventions
#> # A tibble: 3 x 4
#> date_min date_max intervention_label intervention
#> <date> <date> <chr> <chr>
#> 1 2020-06-28 2020-07-09 Seroprevalence study seroprev
#> 2 2020-03-16 2020-06-30 Closed due to COVID-19 closed
#> 3 2020-07-01 2020-09-20 Partially open partial
# interventions %>%
# writexl::write_xlsx("table/02-seroprev-supp-table05.xlsx")
# mobility data -----------------------------------------------------------
mobility <- read_google_region_country(country_iso = "PE")
# mobility %>% count(sub_region_1) %>% print(n=Inf)
# mobility %>% count(country_region,sub_region_1,sub_region_2,metro_area) %>% print(n=Inf)
mobility_lima <- mobility %>%
filter(date<max_analysis_date) %>%
filter(
magrittr::is_in(sub_region_1,c("Metropolitan Municipality of Lima",
"Callao Region"))) %>%
mutate(across(c(sub_region_1,sub_region_2,metro_area),
str_replace_na,replacement = "")) %>%
mutate(subregion=str_c(sub_region_1,"\n",sub_region_2,"\n",metro_area)) %>%
mutate(subregion=str_trim(subregion)) %>%
mutate(subregion=if_else(sub_region_1=="Metropolitan Municipality of Lima",
"Metropolitan Municipality\nof Lima",subregion)) %>%
# count(country_region,subregion)
filter(!(sub_region_1=="Metropolitan Municipality of Lima" &
sub_region_2=="")) %>%
# count(country_region,sub_region_1,sub_region_2,metro_area,subregion)
# pivot_longer()
pivot_longer(cols = -c(country_region_code:date,subregion),
names_to = "field",
names_pattern = "(.+)_percent_change_from_baseline",
values_to = "percent_change_from_baseline")
mobility_lima %>%
mutate(field=str_replace_all(field,"_"," "),
field=str_to_sentence(field)) %>%
ggplot() +
geom_rect(data = interventions,
aes(xmin = date_min, xmax = date_max,
ymin = -Inf, ymax = Inf,
fill =intervention_label),
alpha=0.2) +
geom_hline(aes(yintercept=0),lty=2) +
geom_line(aes(x = date,percent_change_from_baseline, color=subregion)) +
geom_smooth(aes(x = date,percent_change_from_baseline, color=subregion),
span = 0.1) +
facet_wrap(~field) +
colorspace::scale_color_discrete_qualitative() +
scale_x_date(date_breaks = "1 month",date_labels = "%b") +
# theme(legend.position="bottom")
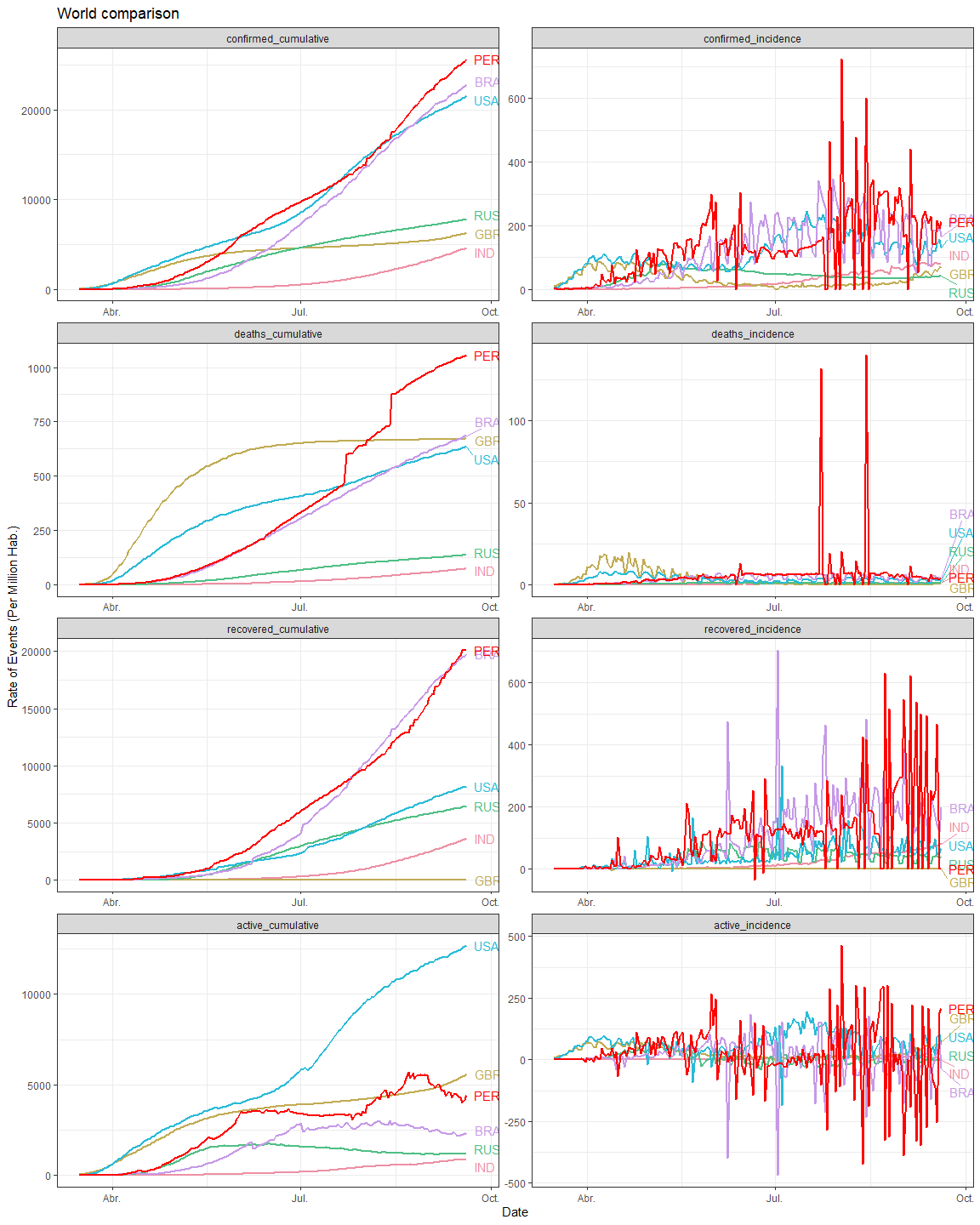
labs(title = "Government interventions and Google mobility reports",
subtitle = "Lima Metropolitan Area and Callao Region, Peru 2020",
x = "Date",
y = "% change from baseline",
fill = "Interventions",
color = "Region")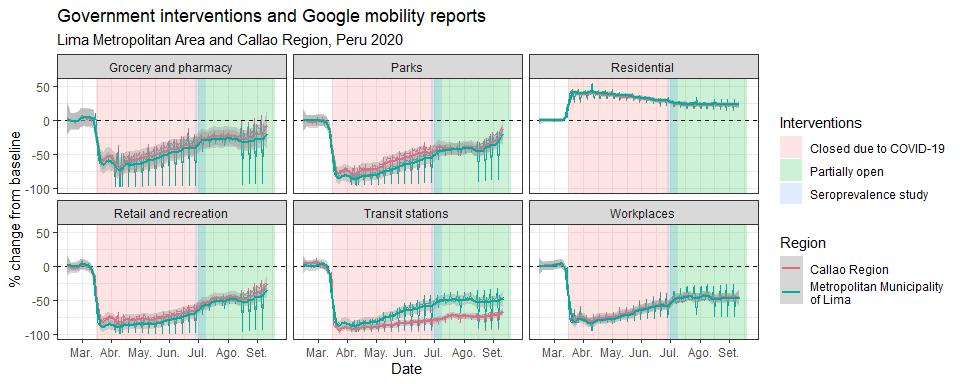
Example 2: Compare event rates among countries
Collect data
library(tidyverse)
library(rnaturalearth)
#world data
world_map = ne_countries(returnclass = "sf")
country_metadata <- world_map %>%
filter(str_detect(string = formal_en,
pattern = "United States|US|Brazil|Russia|Peru|India|United Kingdom")) %>%
as_tibble() %>%
select(contains("name"),pop_est,pop_year,lastcensus,economy,income_grp,iso_a3)
#case data
all <- tibble(country_region=c("US","Brazil","Russia","Peru","India","United Kingdom")) %>%
mutate(counts=map(.x = country_region,.f = jhu_sitrep_all_sources))
all %>%
select(-country_region) %>%
unnest(cols = c(counts)) %>%
select(country_region,source,sum_data) %>%
pivot_wider(names_from = source,values_from = sum_data) %>%
mutate(active=confirmed-deaths-recovered) %>%
pivot_longer(cols = -country_region,names_to = "source",values_to = "sum_data") %>%
mutate(country_region=if_else(country_region=="US","United States",country_region)) %>%
rename(name=country_region) %>%
#join
left_join(country_metadata) %>%
select(name,source,sum_data,pop_est,lastcensus#,economy,income_grp
) %>%
mutate(rate=sum_data/pop_est*1e6) %>%
# group_by(source) %>%
arrange(source,desc(rate)) %>%
knitr::kable() %>%
kableExtra::kable_styling(bootstrap_options = "striped", full_width = F)| name | source | sum_data | pop_est | lastcensus | rate |
|---|---|---|---|---|---|
| United States | active | 3988265 | 313973000 | 2010 | 12702.57315 |
| United Kingdom | active | 348770 | 62262000 | 2011 | 5601.65109 |
| Peru | active | 130616 | 29546963 | 2007 | 4420.62353 |
| Brazil | active | 454815 | 198739269 | 2010 | 2288.50092 |
| Russia | active | 170100 | 140041247 | 2010 | 1214.64214 |
| India | active | 1010824 | 1166079220 | 2011 | 866.85706 |
| Peru | confirmed | 756412 | 29546963 | 2007 | 25600.32989 |
| Brazil | confirmed | 4528240 | 198739269 | 2010 | 22784.82769 |
| United States | confirmed | 6764970 | 313973000 | 2010 | 21546.34316 |
| Russia | confirmed | 1092915 | 140041247 | 2010 | 7804.23642 |
| United Kingdom | confirmed | 392845 | 62262000 | 2011 | 6309.54675 |
| India | confirmed | 5400619 | 1166079220 | 2011 | 4631.43405 |
| Peru | deaths | 31283 | 29546963 | 2007 | 1058.75518 |
| Brazil | deaths | 136532 | 198739269 | 2010 | 686.99055 |
| United Kingdom | deaths | 41848 | 62262000 | 2011 | 672.12746 |
| United States | deaths | 199259 | 313973000 | 2010 | 634.63737 |
| Russia | deaths | 19270 | 140041247 | 2010 | 137.60232 |
| India | deaths | 86752 | 1166079220 | 2011 | 74.39632 |
| Peru | recovered | 594513 | 29546963 | 2007 | 20120.95118 |
| Brazil | recovered | 3936893 | 198739269 | 2010 | 19809.33622 |
| United States | recovered | 2577446 | 313973000 | 2010 | 8209.13263 |
| Russia | recovered | 903545 | 140041247 | 2010 | 6451.99196 |
| India | recovered | 4303043 | 1166079220 | 2011 | 3690.18067 |
| United Kingdom | recovered | 2227 | 62262000 | 2011 | 35.76821 |
Plot trends
all_time <- all %>%
mutate(table=map(.x = counts,.f = jhu_sitrep_all_sources_tidy)) %>%
select(-country_region) %>%
unnest(cols = c(table)) %>%
filter(confirmed_cumulative>0) %>%
#active cases
mutate(active_cumulative=confirmed_cumulative-deaths_cumulative-recovered_cumulative,
active_incidence=confirmed_incidence-deaths_incidence-recovered_incidence) %>%
mutate(country_region=if_else(country_region=="US","United States",
country_region)) %>%
rename(name=country_region) %>% #glimpse()
select(-counts) %>%
pivot_longer(
# cols = confirmed_cumulative:recovered_incidence,
cols = confirmed_cumulative:active_incidence,
names_to = "indicator",values_to = "value") %>%
#join
left_join(country_metadata) %>%
select(iso_a3,name:value,pop_est,lastcensus) %>%
mutate(rate=value/pop_est*1e6) %>% #count(indicator)
mutate(name=fct_reorder(.f = name,.x = rate,.fun = max)) %>%
mutate(indicator=fct_relevel(indicator,
"confirmed_cumulative",
"confirmed_incidence",
"deaths_cumulative",
"deaths_incidence",
"recovered_cumulative",
"recovered_incidence",
"active_cumulative",
"active_incidence"
)) %>%
filter(date>lubridate::ymd(20200315)) %>%
mutate(look=if_else(iso_a3=="PER","PER","NOT"))
# group_by(name,indicator) %>%
# mutate(label=max(rate)) %>%
# ungroup()
all_time %>%
ggplot(aes(x = date,y = rate,colour = name)) +
geom_line(data = all_time %>% filter(look!="PER"),
lwd=1) +
facet_wrap(vars(indicator),scales = "free",ncol = 2) +
# scale_colour_viridis_d(option = "A")
colorspace::scale_color_discrete_qualitative(palette = "Set 2",
guide = 'none') +
geom_line(data = all_time %>% filter(look=="PER"),
color="red",lwd=1) +
xlim(c(lubridate::ymd(20200315),Sys.Date()+5)) +
ggrepel::geom_text_repel(data = all_time %>%
group_by(name,indicator) %>%
filter(date==max(date)) %>%
ungroup() %>%
filter(look!="PER"),
aes(label = iso_a3),
xlim = c(Sys.Date()+1, NA),
hjust = 1) +
ggrepel::geom_text_repel(data = all_time %>%
group_by(name,indicator) %>%
filter(date==max(date)) %>%
ungroup() %>%
filter(look=="PER"),
color="red",
aes(label = iso_a3),
xlim = c(Sys.Date()+1, NA),
hjust = 1) +
# scale_y_log10(
# breaks = 10^(-10:10),
# minor_breaks = rep(1:9, 21)*(10^rep(-10:10, each=9))
# ) +
# # scale_y_log10(
# # # breaks = scales::pretty_breaks(n = 10)
# # # breaks = scales::trans_breaks("log10", function(x) 10^x),
# # # labels = scales::trans_format("log10", scales::math_format(10^.x))
# # ) +
# # coord_trans(y="log10") +
# annotation_logticks(sides = "lb") +
labs(title = "World comparison",
x = "Date",y="Rate of Events (Per Million Hab.)")